Introduction
Cytokines play an important role in modulation of the immune response. They are produced by immune cells upon stimulation. By binding to specific receptors cytokines can either up-regulate activation, proliferation and differentiation of target cells, mediate or regulate immune reactions, inhibit the growth of cells, act cytotoxic, induce or inhibit the production of other cytokines. Cytokines are divided in two groups according to the function: anti-inflammatory and pro-inflammatory cytokines. Lack of balance between pro- and anti-inflammatory cytokines disables proper function of immune system. In recent years, many researchers have noticed that differences in cytokine levels (high or low) are associated with certain allelic variants of cytokine genes. These polymorphisms might play an important role in the pathophysiology of various diseases.
Interleukin 10 (IL-10) is an important pleiotropic immunoregulatory cytokine mainly secreted by macrophages, but also by T helper 1 (Th1) and Th2 lymphocytes, dendritic cells, cytotoxic T cells, B lymphocytes, monocytes and mast cells. Some studies have shown that it can be produced even by human carcinoma cell lines (1,2). IL-10 activity is mediated by the IL-10 receptor (IL-10R) which is a member of the class II cytokine receptor family. IL-10 inhibits the capacity of monocytes and macrophages to present antigen to T cells via an inhibitory effect on expression of major histocompatibility complex (MHC) class II, costimulatory molecules such as CD80 (B7.1) and CD86 (B7.2) and therefore downregulates the expression of IL-1, IL-6, IL-8, IL-12 and tumor necrosis factor – alpha (TNF-α). In B cells, IL-10 prevents apoptosis, enhances cell proliferation and has a role in immunoglobulin (Ig) class switch.
The IL-10 gene is located on chromosome 1 at 1q31-32, spans about 4.7 kb and contains four introns and five exons (3). There are many genetic variants of IL-10 gene. However, the most studied are two dinucleotide repeats (microsatellites), IL10.G and IL10.R, located 1.2 kb and 4 kb upstream of the transcription start site (4,5) and three single nucleotide polymorphisms (SNPs) -1082(G/A), -819(C/T) and -592(C/A) (6) which form three predominant haplotypes (GCC, ACC, ATA). Although endogenous and exogenous factors stimulate cells to produce IL-10, its secretion also depends on IL10.R, IL10.G and SNP polymorphisms in promoter region. A summary of all IL-10 polymorphisms and haplotypes associated with diseases and their links to expression patterns are shown in Table 1.
Table 1. Summary of IL-10 polymorphisms associated with diseases and their links to expression patterns.
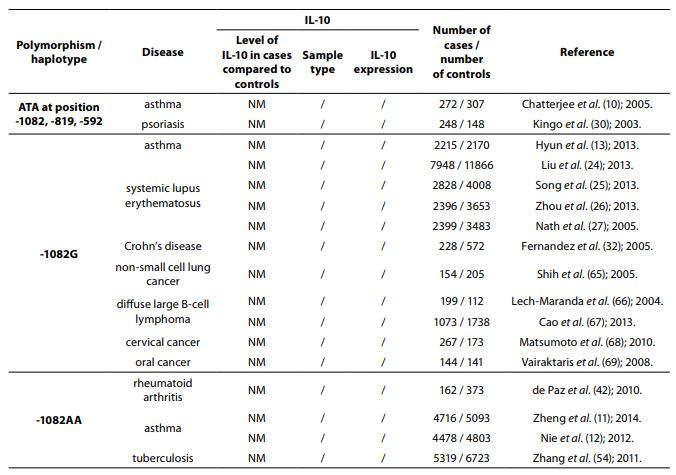
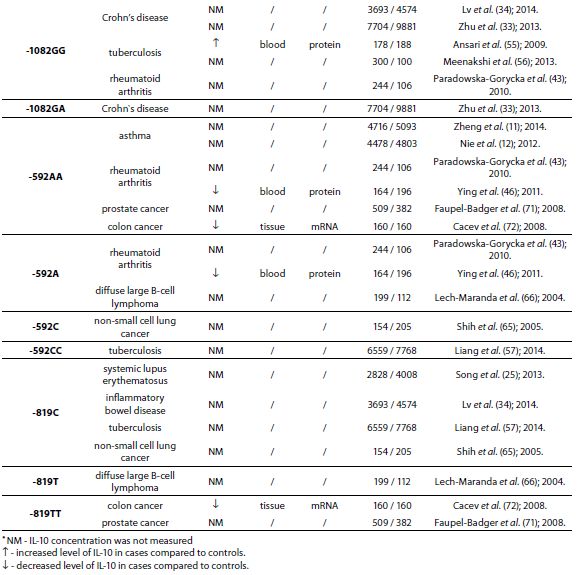
The pleiotropic role of IL-10 in immune system regulation coupled with polymorphic regulation of its expression, presents additional challenges for interpretation, especially in the context of disease pathophysiology. The aim of this review was to present the most studied associations of IL-10 polymorphisms with susceptibility to selected diseases and their severity. A literature search was conducted using PubMed and SCOPUS from January 2000 through to April 2014. Additional literature references were found by reviewing reference lists. Search was first performed using the search term “Interleukin 10 polymorphism” to discover which IL-10 polymorphisms and diseases were studied the most. Those were three single nucleotide polymorphisms (SNPs): 1082(G/A), -819(C/T) and -592(C/A) in the promoter region which form three predominant haplotypes (GCC, ACC, ATA). The most studied diseases were asthma, systemic lupus erythematosus, psoriasis, inflammatory bowel disease, rheumatoid arthritis, tuberculosis and neoplasms. Further search was referred only to these diseases. Inclusion criteria for the review were publications limited to humans, well design case-control studies (sample size > 100, well-defined group of cases and controls, detailed methods, P < 0.05), meta-analysis (with stratification by age or ethnic origin if possible). No age limits were set. Power analysis has not been performed in any of the studies included in this review. All data are presented systematically and summarized in tables sorted according to the type of illness (IL-10 polymorphism / haplotype associated with the disease, frequency of polymorphism / haplotype in cases compared to controls, population represented in the study).
IL-10 in autoimmune and inflammatory diseases
Asthma
Asthma is a complex disorder in which both genetic and environmental risk factors play an important role in pathogenesis of the disease. Pro-inflammatory cytokines produced by Th2 cells have an important role in the chronic inflammation in asthma since they are responsible for IgE dysregulation (IL-4, IL-13), eosinophilia (IL-5, granulocyte-macrophage–colony stimulating factor (GM-CSF), IL-3), mast cell proliferation (IL-3, IL-9), IgE class switching (IL-4, IL-13) and hypersecretion of mucus and regulation of airway hypersensitivity (IL-13). Therefore, asthma is considered to be a Th2 disease (7). The intrinsic physiologic mechanism for inhibiting pro-inflammatory cytokine synthesis is cytokine IL-10 (8). In healthy lungs the main sources of IL-10 are alveolar macrophages and circulating monocytes (9). In bronchoalveolar lavage (BAL) alveolar macrophages represent more than 80% of the cells present, while Th1 and Th2 lymphocytes, cytotoxic T cells, B lymphocytes and mast cells represent less than 10% of the cells (8).
An inverse association between severity of asthma and IL-10 concentration was shown by Borish et al. (8) who measured concentration of IL-10 in BAL in normal and asthmatic subjects using IL-10 enzyme-linked immunosorbent assay (ELISA). In BAL fluid of asthmatic patients IL-10 concentration was lower than in healthy controls (P < 0.01). Absence of IL-10 in asthma causes the continued secretion of pro-inflammatory cytokines such as IL-6, IL-5, IL-4, TNF-α, GM-CSF and IL-1 which contribute to asthmatic airway inflammation.
Many candidate genes and SNPs have been studied and found out to be associated with asthma (Table 2).
Table 2. Association of IL-10 polymorphism and haplotype with asthma.
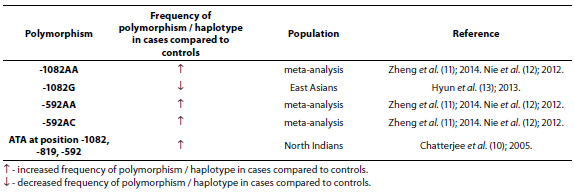
Chattarjee et al. (10), using both case-control and family studies, showed that haplotype ATA at position -1082, -819, -592 was present more in asthmatics (N = 272) than in controls (N = 307) (P = 0.009) while haplotype ATC at position -1082, -819, -592 was more present in controls than in asthmatics (P = 0.012) in North Indians. IL-10 meta-analysis involving 4716 asthmatic patients and 5093 controls showed the association of -1082AA, -592AC and -592AA genotypes with asthma susceptibility in Asian adults (11). This association was also shown in meta-analysis involving 4478 asthmatic patients and 4803 controls from different populations (12). Eleven studies involving 2215 asthma patients and 2170 controls were included in a meta-analysis performed by Hyun et al. (13). Stratification by ethnicity indicated an association between the IL-10 -1082G allele and asthma in East Asians (P = 0.02). Stratification by age indicated an association between the IL-10 -1082G allele and asthma in adults (P = 0.02).
Systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterized by B lymphocyte hyperactivity and production of autoantibodies directed against double-stranded DNA, dysfunction of antigen-presenting cells (APC) and T lymphocytes. Increased production and decreased clearance of immune complexes leads to immune complex deposition in tissue and damage to multiple organ systems. IL-10 has the ability to induce autoantibody production by B lymphocytes suggesting that IL-10 plays an important role in the pathogenesis of SLE (14).
The major sources of IL-10 in patients with SLE are B cells and monocytes. IL-10 overproduction by B lymphocytes and monocytes was described for the first time by Llorente et al. (15). This observation was also confirmed by several other studies (16,17). Abnormal production of autoantibodies by B lymphocytes in patients with SLE is IL-10 dependent and all related studies demonstrated that there is a positive correlation of serum IL-10 levels with disease activity (18-20). Interferons that are produced early in disease, as part of the immune response, have capacity to change IL-10 function from anti- to pro-inflammatory, so in this context IL-10 contributes to inflammation (21). In addition, immune complexes, that are produced in SLE, acting through Fc gamma receptor II (FcgRII) stimulate IL-10 production from peripheral blood mononuclear cells (PBMC), thus perpetuating the pathological cycle. Rönnelid et al. (22) showed a significant increase in IL-10 production in cell cultures incubated with SLE sera, in comparison to cell cultures incubated with control sera. This effect provides a possible explanation for the enhanced production of IL-10 in patients with SLE, which leads to B cell hyperactivity, autoantibody production, immune complexes production, PBMC stimulation and again production of IL-10. Creation of this cycle leads to increased deposition of immune complexes in tissues and SLE associated pathology.
Numerous studies have investigated the association between the -1082(G/A), -819(C/T) and -592(C/A) polymorphisms and susceptibility to SLE, but the results were inconsistent. Therefore, a large number of meta-analysis in the last few years have been performed to assess the association between SLE and the IL-10 polymorphisms (Table 3).
Table 3. Association of IL-10 polymorphism with systemic lupus erythematosus.

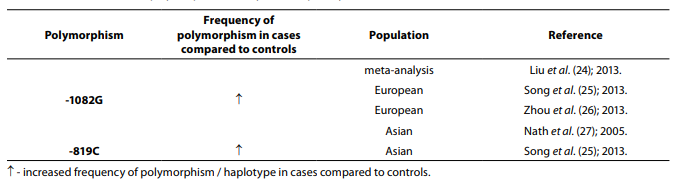
Twelve studies including 1765 cases and 2444 controls were included in a meta-analysis performed by Wang et al. (23). The results indicated that there is a lack of association between the haplotype GCC at position -1082, -819, -592 and SLE risk, but another meta-analysis that included 7948 cases and 11866 controls has shown that the haplotype GCC at position -1082, -819, -592 is associated with SLE susceptibility same as -1082G allele (24). In meta-analysis involving 2828 SLE patients and 4008 controls (25), stratification by ethnicity indicated an association between the GCC haplotype and SLE in Europeans, IL-10 -819C allele and SLE in Asians, while an association between the IL-10 -1082G allele and SLE in Europeans was also indicated in meta-analysis (2396 cases and 3653 controls) performed by Zhou et al. (26). Meta-analysis that included 2399 SLE patients and 3483 controls showed that -1082G allele is associated with SLE in Asians (27).
Psoriasis
Psoriasis is a cutaneous disorder characterized by overexpression of pro-inflammatory Th1 cytokines (IL-2, interferon-gamma (IFN-γ), TNF-α) while the level of expression of anti-inflammatory cytokines (IL-4, IL-10) is low and insufficient to counterbalance pro-inflammatory effects. Low levels of IL-10 were observed in many studies (28,29) and it is believed that it plays a key role in the pathology and in the clinical course of psoriasis.
Polymorphisms of IL-10 promoter region in psoriasis were frequently investigated. In a study that involved 248 patients with plaque type of psoriasis and 148 controls it was shown that ATA haplotype has a role in determining severity and course of plaque type of psoriasis since it is associated with persistent eruption (P < 0.01) (30). A meta-analysis involving Asian psoriasis patients (N = 1018) and controls (N = 1186) found significant association between psoriasis and the IL-10 -1082G allele (P = 0.011) (31).
Inflammatory bowel disease
Inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC), is a chronic relapsing disease characterized by inflammation of gastrointestinal tract, which leads to destruction of the mucosa. An important element in pathogenesis of IBD is disregulation of the intestinal immune system. In the normal intestinal immune system the balance of pro- and anti-inflammatory cytokines is essential for gut homeostasis. However, in IBD peripheral blood monocytes, intestinal monocytes and polymorphonuclear neutrophil granulocytes produce high amounts of pro-inflammatory cytokines (IL-6, TNF-α, IL-1β, IL-8) which leads to differentiation or proliferation of a variety of cells and tissue damage.
Genetic factors were also found to be associated with the development of UC and CD (Table 4).
Table 4. Association of IL-10 polymorphism with inflammatory bowel disease.
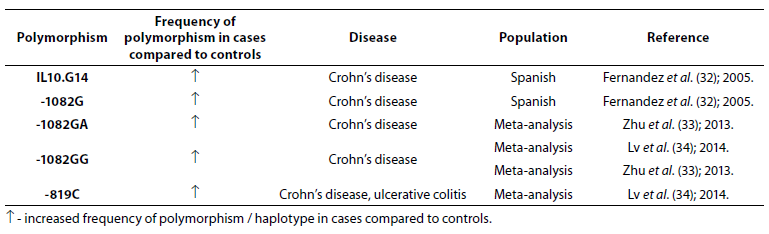
Frequencies of IL10.G14 microsatellite allele and IL-10 -1082G allele were higher in patients with CD (N = 228) compared to patients with UC (N = 242) and controls (N = 572) (32). Zhu et al. (33) performed meta-analysis (7704 IBD patients and 9881 controls) to estimate the association between -1082A/G polymorphism in the IL-10 gene and IBD susceptibility. The results demonstrated association between heterozygote genotypes -1082GA and -1082GG and CD. Another meta-analysis (3693 cases and 4574 controls) showed association of -1082GG genotype and CD and -819C and IBD (34).
Rheumatoid arthritis
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease that is characterized with chronic synovitis, which often leads to joint destruction. Expression of pro- and anti-inflammatory cytokines in the synovial membrane of the inflamed joint is altered. Pro-inflammatory cytokines, TNF-α and IL-1, are overproduced by macrophages, synovial fibroblasts and neutrophils and play an important role in the process of chronic inflammation and joint destruction.
Synovial fluid from patients with RA also contains detectable levels of anti-inflammatory cytokine IL-10 but they are insufficient to counterbalance the effect of pro-inflammatory cytokines (35). The cellular sources of IL-10 in the synovial tissue are macrophages and T cells (35). In the presence of TNF-α and IL-1 expression of IL-10R on surface of dendritic cells in synovial fluid is low (36). Also, chronic exposure of the pro-inflammatory cytokines (TNF-α and IL-1) modulates the IL-10 signalling and bioactivity in synovial macrophages during chronic inflammation (37). RA patients showed significantly reduced IL-10 levels in serum in comparison to healthy donors, suggesting that IL-10 synthesis is depressed in RA (38). In addition, patients with advanced RA had the lowest values of IL-10 in serum (39). On the contrary, some studies have shown that concentration of IL-10 in sera of patients with RA is higher as compared to controls (40,41). Polymorphisms within the IL-10 gene promoter are associated with RA (Table 5).
Table 5. Association of IL-10 polymorphism and haplotype with rheumatoid arthritis.
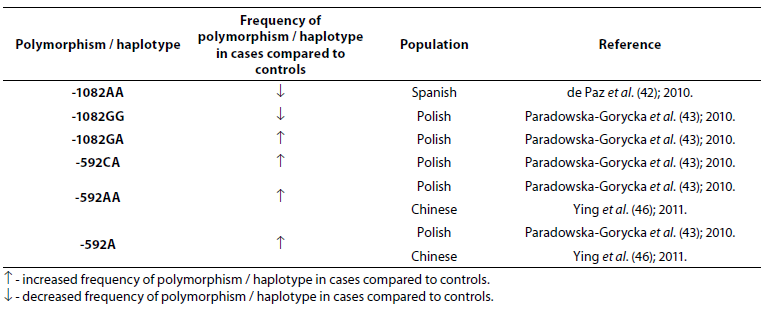
Frequency of genotype -1082AA was lower (P = 0.006) in RA patients (N = 162) when compared to the control group (N = 373) (42). In study performed by Paradowska-Gorycka et al. (43) it was found that frequency of genotype -1082GG was lower (P = 0.0001) while frequency of genotype -1082GA was higher (P = 0.009) in RA patients (N = 244) comparing to the control group (N = 106). A total of 1480 cases and 1413 controls in 10 case-control studies were included in meta-analysis performed by Zhang et al. (44). The results indicated that people with IL-10 -1082GG and -1082GA genotype had a 25% lower risk of RA, when compared with the people with IL-10 -1082AA genotype. Meta-analysis (2647 RA patients and 3383 controls) showed association of the IL-10 -592C allele and the IL-10 -592CC genotype with RA (45). In Polish (43) and Chinese (46) RA patients frequency of -592AA genotype was significantly higher compared to controls, same as frequency of genotype -592CA and allele -592A, suggesting that allele A is associated with the lower expression of the IL-10 protein.
Tuberculosis
Tuberculosis (TBC) is caused by Mycobacterium tuberculosis, an intracellular pathogen, which primarily enters the human organism through respiratory tract and is phagocytised by macrophages and dendritic cells. Phagocytic cells undergo activation and maturation, which increases their potential to stimulate T cells. Cytokines released by activated T cells regulate differentiation, proliferation and effector functions of phagocytic cells with a purpose of removing intracellular pathogen and limiting disease. Th1 cytokines are produced during early stage of the infection while Th2 cytokines are produced during later stages. Balance between Th1 and Th2 cytokines plays an important role in regulating Mycobacterium tuberculosis infection. Components of the Mycobacterium tuberculosis cell wall stimulate production of IL-10 by macrophages, but with a progression of inflammation, T cells become a major source of IL-10 production.
Studies have shown that IL-10 is associated with progression of disease caused by Mycobacterium tuberculosis (47,48). Increased IL-10 down-regulates secretion of important anti-mycobacterial molecules: TNF-α and nitric oxide, expression of costimulatory molecules and major histocompatibility complex (MHC) class II by macrophages and IFN-γ production by T cells (49,50). These findings suggest that IL-10 is associated with promotion of pathological processes and Mycobacterium tuberculosis survival inside host macrophages during infection.
High concentration of IL-10 has been found in sera (51) and lungs (52) of patients with tuberculosis. Neutralization of endogenous IL-10 enhanced T cell IFN-γ production, expression of costimulatory molecules and accelerated the clearance of mycobacteria. Association of IL-10 polymorphism with tuberculosis is shown in Table 6.
Table 6. Association of IL-10 polymorphism and haplotype with tuberculosis.
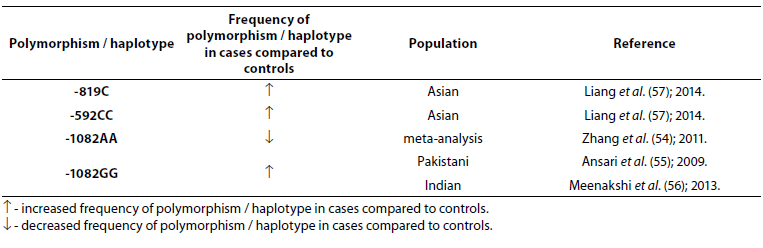
There were no statistically significant differences observed between patients with TBC and controls for the frequency of IL-10 -1082 alleles or genotypes (P > 0.05); however, a statistically significant difference in the frequency of IL-10 -1082GG genotype was found between patients with pulmonary and extrapulmonary TBC (P = 0.003) (53). Genotype -1082AA (low IL-10 producer) is associated with resistance to pulmonary tuberculosis (54) while genotype -1082GG (high IL-10 producer) is associated with the risk of developing tuberculosis (55,56). Meta-analysis that included 31 studies with 6559 cases and 7768 controls showed that three polymorphisms (-1082G/A, -819T/C and -592A/C) in the IL-10 gene were not associated with the risk of TBC in general population, but in the subgroup analysis by ethnicity, IL-10 -1082AA and -1082AG genotypes were associated with the TBC risk in Europeans and Americans, while IL-10 -819C allele and -592CC and -592AC genotypes were significantly associated with the TBC risk in Asians (57).
IL-10 in melanoma and other malignancies
Cancer is a multifactorial disease, resulting from complex interactions between genetic and environmental factors. Cytokines also have an important role in cancer. They can either inhibit tumour development and progression if they are released in response to inflammation or they can induce tumour invasion and metastasis if they inhibit apoptosis or promote growth. Since IL-10 has pleiotropic effects, its role and polymorphisms have been widely studied in different types of cancer. The role of IL-10 in cancer is best described in development and progression of melanomas.
Kriiger-Krasagakes et al. (58) investigated the expression of IL-10 mRNA in tissue specimens of primary malignant melanomas and melanoma metastases as compared with normal skin. Their results showed that IL-10 mRNA was found in primary tumours tissues and metastases but not in normal skin. However, they could not distinguish which cells in the tumour tissue are producing IL-10 mRNA, infiltrating cells (B cells, T cells and monocytes) or melanoma cells. Sato et al. (59) showed by cell separation experiments and intracellular staining that the melanoma cells themselves are the major source of IL-10. They did not exclude the possibility that tumour-associated leukocytes also produce IL-10 but suggested that their contribution would be minor.
IL-10 production by tumour cells increases as primary melanomas progress from melanoma in situ to invasive melanoma and metastatic melanoma (60). It is believed that IL-10 may have different roles during tumour development and progression. Melanoma cells express IL-10R (61) so IL-10 acts as an autocrine growth factor. In the presence of IL-10 the proliferation of melanoma cells was increased, the cells grew faster and the survival period of cells was prolonged. Melanoma cells also express gangliosides such as GD3 and GM3, which are important for tumour growth and metastasis. CD1 molecules, which are expressed by APC such as dendritic cells, recognize those gangliosides and destruct the cells. However, melanoma cells, by secreting IL-10, down-regulate CD1 molecules on infiltrating dendritic cells and escape the immune response directed against the tumour (62). IL-10 also down-regulates human leukocyte antigen (HLA) class I, HLA class II and intercellular adhesion molecule 1 (ICAM-1) on human melanoma cells and therefore also the T-cell response. Apart from this immune-evading mechanism, it was demonstrated that IL-10 shows an anti-tumour effect via inhibition of angiogenesis (63), so its function in melanomas remains complex and still unclear. Presence of low producing ATA haplotype at position -1082, -819, -592 is associated with the susceptibility to melanoma (64).
Association of IL-10 polymorphisms and other malignancies are shown in Table 7.
Table 7. Association of IL-10 polymorphism and haplotype with other malignancies.
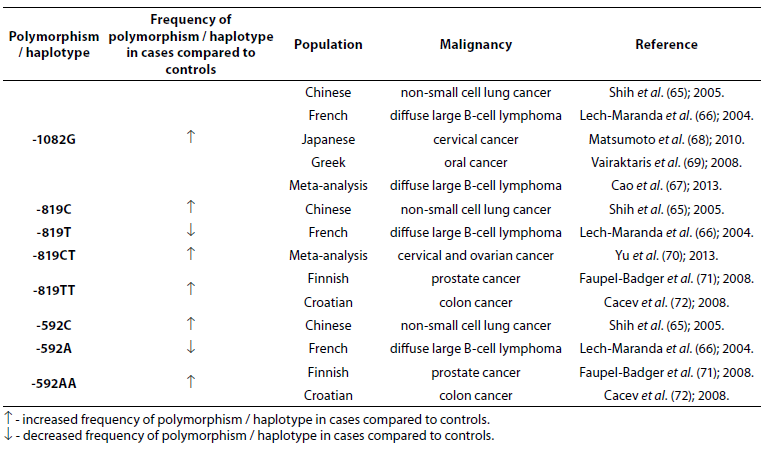
IL-10 -1082G allele is associated with numerous other malignancies: non-small cell lung cancer (NSCLC) (65), diffuse large B-cell lymphoma (DLBCL) (66,67), cervical (68) and oral (69) cancer. In Chinese population alleles -819C and -592C were significantly associated with the occurrence of NSCLC (65). The frequencies of -819T and -592A alleles were lower in French patients (N = 199) with DLBCL compared to controls (N = 112) (66). Yu et al. (70) in a meta-analysis, based on 73 studies that included 15942 cases and 22336 controls, found that -819CT genotype is a risk factor for cervical and ovarian cancer. Prostate cancer in Finish (71) and colon cancer in Croatian populations (72) are associated with low IL-10 mRNA expression genotypes -819TT and -592AA. No significant differences in allele frequency or genotype distribution were observed for any of the IL-10 SNPs between patients with prostate cancer (N = 262) and control subjects (N = 270). However, significantly higher frequencies of -1082G (P = 0.005), -819C (P = 0.043) and -592C (P = 0.043) allele and GCC haplotype (P = 0.008) were observed in early stage patients in comparison to advanced prostate cancer patients (73).
Conclusion
Production of IL-10 is stimulated by various endogenous and exogenous factors, but certain allelic variants of IL-10 gene are also associated with differences in IL-10 expression. Genetic association studies are performed to determine whether a genetic variant is associated with the disease. IL-10 has been shown to be a candidate gene in the pathophysiologic mechanism of autoimmune diseases, inflammatory diseases and some neoplasms since it regulates both cellular and humoral immunity. Three SNPs (-1082(G/A), -819(C/T), -592(C/A)) in the promoter region of the IL10 gene have been shown to alter IL-10 mRNA and protein levels. High expression of allele -1082G in patients compared to controls is associated with SLE, CD and neoplasms (non-small cell lung cancer, diffuse large B-cell lymphoma, cervical and oral cancer) and promotes development of pathological processes in these diseases. Genotype -1082AA is more prevalent in asthma, while in TBC and Crohn’s disease genotype -1082GG has higher frequency in cases compared to controls. Genotype -819TT is associated with low level of IL-10 in patients with colon cancer. In the presence of low IL-10 production allele -592A and genotype -592AA in RA, expression of anti-inflammatory cytokine IL-10 is low and insufficient to counterbalance the effect of pro-inflammatory cytokines. In patients with asthma, prostate and colon cancer frequency of low IL-10 production genotype -592AA is higher compared to controls. However, it is necessary to perform additional studies to fully understand the association of IL-10 polymorphisms with disease predisposition and underlying pathological processes.
Both overexpression (SLE, tuberculosis) as well as IL-10 deficiency (inflammatory bowel disease, psoriasis, asthma, rheumatoid arthritis) are likely to have a pathophysiological significance. Therefore, neutralization of the cytokine could be a promising approach to treat diseases from the first group, whereas application of IL-10 itself could be helpful for diseases from the second group. The role of IL-10 as a therapeutic agent has been described in other reviews (74,75,76,77).


