Introduction
Improvement in patient safety through error reduction is rightfully receiving a great deal of focus in medicine including pathology as it plays a pivotal role in medical decision making (1). Published data states that up to 80% of patient care decisions are based on pathology data (2), and in some cases pathology data is the sole information used in the clinical decision process. With the continuously increasing number of tests, pathology will play even greater role thus laboratorians will need to become actively integrated in the medical care team and aid doctors with interpretation of laboratory results to reduce medical errors (3). Various estimates have been provided on the sources and errors rates in chemical pathology. It is well accepted of the three phases of the pathology testing cycle: the analytical phase is the least error prone while the pre-analytical is the most error prone (4,5). A lot of these errors can be linked to the analytical sample integrity of which lipaemia is a contributor. Even though lipaemic samples are not frequently encountered, these samples continue to provide challenges for a lot of laboratories, more specifically in establishing good practices for processing lipaemic samples and providing accurate results for best patient outcomes (6).
The lipid induced interferences can originate naturally in the patient sample as a result of health-related factors e.g. dietary, diabetes mellitus, alcohol abuse, hypothyroidism, pancreatitis, drug-induced such as oral contraceptives etc (7), from lipid emulsions used for nutritional need in critically ill patients such as newborns (8) and surgical patients (9), and in treatment of lipophilic drug overdoses (10). A review of literature shows it is sparse on lipaemic studies, and there was no published data indicating the extent of the problem. Unlike haemolysis and bilirubin studies lipaemic studies are often left in the too hard basket. This is because of the lack of readily available standardized materials like those used for bilirubin or haemoglobin to mimic many of the properties of native lipaemic samples to produce the lipaemic interferences complicates the evaluation of lipaemia (11). Lipaemia presents other unique problems which include differences in the final lipid composition from patient to patient, the volume of single native sample normally being too small to do various studies, and samples not being able to be frozen for future studies. An additional complication is increasing hyperlipidaemia is associated with increased haemolysis “strawberry milk appearance”, possibly the result of increased erythrocyte membrane fragility induced by alterations in membrane lipid content (12). Hence, artificial compounds i.e. lipid emulsion e.g. Intralipid are used to perform lipaemic index and analyte interference studies by manufacturers and laboratories. Lipid emulsions consist predominantly of small, relatively dense, phospholipid-rich liposomes and triglyceride-rich artificial chylomicrons and no complex mixture of lipoproteins (13). Thus, because of these composition differences there is poor association between lipaemic index and triglyceride concentrations (14,15). Triglycerides are usually the most abundant lipids in lipaemia.
Sample turbidity from lipaemia affects most significantly the photometric assays (end point, rate, nephelometric or turbidimetric) due to increased light scatter and absorption of the light by the lipids (mainly chylomicrons and very low density lipoproteins) (16), and this includes coagulation assays (17). Furthermore, the extent of the effect will be dependent on the sampling mechanism, the lipid composition of the sample and whether the sample is well mixed to minimise chylomicrons settling out at the top of the sample. No blanking method can overcome all the interferences from lipaemia. Such samples have the potential to equally affect all assays including immunoassays by binding of lipophilic compounds (e.g. drugs) and blocking/masking the binding sites with reagents to produce timely and measurable end points (6). The increase in the non-aqueous phase leads to volume displacement errors or exclusion effect with methods that do not measure activity of the analyte (e.g. indirect ion selective electrodes used exclusively on laboratory analysers) (18). Unlike haemolysis the interference of lipaemia can be reduced to eliminate the interference by removal of most of the interfering lipids. Methods for removal include ultracentrifugation (the gold standard), high speed centrifugation and lipid clearing agents e.g. LipoClear and n-hexane. The CLSI Interference testing guidelines recommend clarifying the sample using ultracentrifuge (19) but not many laboratories are equipped with such centrifuges. However, just about all laboratories (large or small) possess high speed micro-centrifuges. We are aware many laboratories use high speed micro-centrifuges for lipid reduction without having the effectiveness of the procedure confirmed. One such study found the recovery from high speed centrifugation (10.000 x g) of samples was unacceptable for total bilirubin and CRP (20). The same study found LipoClear treatment produced unacceptable recovery with GGT, CK-MB, total cholesterol, HDL-cholesterol and CRP (20). In this study we compare a procedure with one such high speed micro-centrifuge with an ultracentrifuge for effectiveness in lipid reduction and suitability for routine use in native lipaemic patient samples using a set of analytes we have observed over years to be most affected by lipaemia on our Beckman DxC800 analysers.
Methods and materials
Samples
The samples used in this study were received in several laboratories for routine analysis. The samples were selected for the study based on the triglyceride level alone in order to cover the highest triglyceride concentration range possible. The samples with similar triglyceride concentration levels were pooled to provide sufficient volume for analysis: neat, post ultracentrifugation and post high speed centrifugation. All samples were native serum, and no samples from patients receiving nutritional lipid supplement (total parenteral nutrition – TPN) were used in preparing any of the pools. The samples of interest were stored at 2-8 °C and analyzed within two weeks of commencing the pooling to minimise any deterioration in sample quality. To our observation over many years TPN samples containing lipid emulsions do not exceed 20 mmol/L triglyceride concentrations, most times they are < 10 mmol/L. In total 10 such sample pools were analysed for sodium (indirect ion selective electrode), creatinine (Jaffe), urate (uricase)), total protein (biuret), lactate dehydrogenase (LD) (lactate to piruvate), magnesium (calgamite), cholesterol (cholesterol esterase) and triglycerides (glycerophosphate oxidase).
Samples were mixed by multiple inversions before analysis to eliminate the chylomicrons and other large molecules from forming a layer at the top of the sample. The sampling probes of analysers only descend 2-3 mm below the meniscus to aspirate the test aliquot, and if the aliquot is aspirated from a sample that is not well mixed (lipid rich component) the interference will be exaggerated (6).
Equipment
Each sample pool was split into three aliquots: neat, ultracentrifugation and high speed centrifugation. The ultracentrifugation was performed in a Beckman Coulter Airfuge for 15 minutes at 107.000 x g. The lipid reduced aliquot was then carefully transferred without disturbing the lipid layer into an appropriate aliquot tube for analysis. The high speed centrifugation was performed in a Heraeus Biofuge Primo micro-centrifuge for 15 minutes at 21.885 x g in two Micro tubes (1.5 mL Ref 72.690.001 Sarstedt). The lipid cleared sample, infranatant (lower part of the sample) was transferred into clean Micro tubes using glass pipettes by first expelling the air out of the pipette and then immersing it to the bottom of the Micro tube to aspirate ~2/3 of the sample/infranatant and prevent any bubbles being released during the pipette descent through the supernatant and re-mixing the sample. Furthermore, aspiration was carried out slowly to minimise intermixing between the lipid poor infranatant and lipid rich supernatant layers. This procedure was repeated post re-centrifugation, and ~2/3 of the clear infranatant was aspirated from the two Micro tubes into an appropriate aliquot tube for analysis.
The neat, ultracentrifuged and high speed centrifuged aliquots were analysed on a Beckman DxC800 general chemistry analyser (Beckman Coulter, Fullerton, CA, USA) at the same time on the same analyser in the same rack and the same order for each sample pool. The total analytical imprecision expressed by the between-run coefficient of variation (CV) for the analytes was as follows: Na+ (1.0% at 132 mmol/L and 0.9% at 150 mmol/L), creatinine (4.9% at 68 μmol/L and 1.8% at 491 μmol/L), urate (1.9% at 0.23 mmol/L and 1.5% at 0.49 mmol/L), total protein (1.8% at 41 g/L and 1.6% at 67 g/L), LD (2.7% at 156 U/L and 2.3% at 432 U/L), Mg (2.5% at 0.84 mmol/L and 2.3% at 1.63 mmol/L), cholesterol (1.7% at 3.0 mmol/L and 2.3% at 6.5 mmol/L) and triglycerides (3.6% at 1.0 mmol/L, 2.7% at 2.0 mmol/L and 0.5% at 8.3 mmol/L).
Statistical analysis
The mean and standard deviation were calculated for each analyte for each of the three aliquots. From the imprecision data the least significant change (LSC) (2.77 x √analytical coefficient of variation) was calculated to determine the maximum allowable difference in test results between the different aliquots.
Results
A search of our laboratory information systems showed very small percentage of all the samples requested for lipid analysis had elevated lipids. Currently only ~0.7% and 0.2% of samples requested for lipid studies have triglyceride concentrations > 10.0 mmol/L and > 20.0 mmol/L respectively in our tertiary level referral hospital.
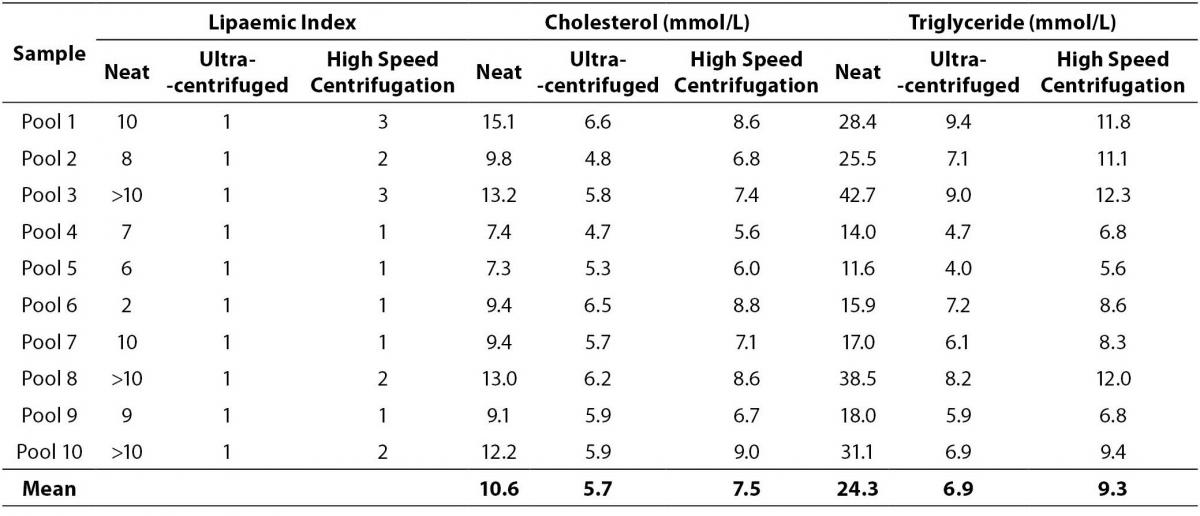
The procedure evaluated in this study using the Biofuge Primo high speed micro-centrifuge showed it is capable of providing a sample with significantly decreased lipid levels close to those obtained by ultracentrifugation (Table 1). The mean differences from the neat aliquot for the ultracentrifuged and high speed centrifuged sample pools were: cholesterol 4.9 mmol/L and 3.1 mmol/L; and triglycerides 17.4 mmol/L and 15.0 mmol/L respectively. The data confirms when several re-centrifugation steps are performed high speed centrifugation is almost as effective as ultracentrifugation in lipid reduction. Although pools one, two, three and eight had triglyceride levels > 10.0 mmol/L, no analyte results were suppressed due to interference. The removal of the majority of the chylomicrons during the high speed centrifugation is likely to have removed of the interference.
Table 1. Comparison of cholesterol and triglycerides concentrations decrease post ultracentrifugation and high speed centrifugation.
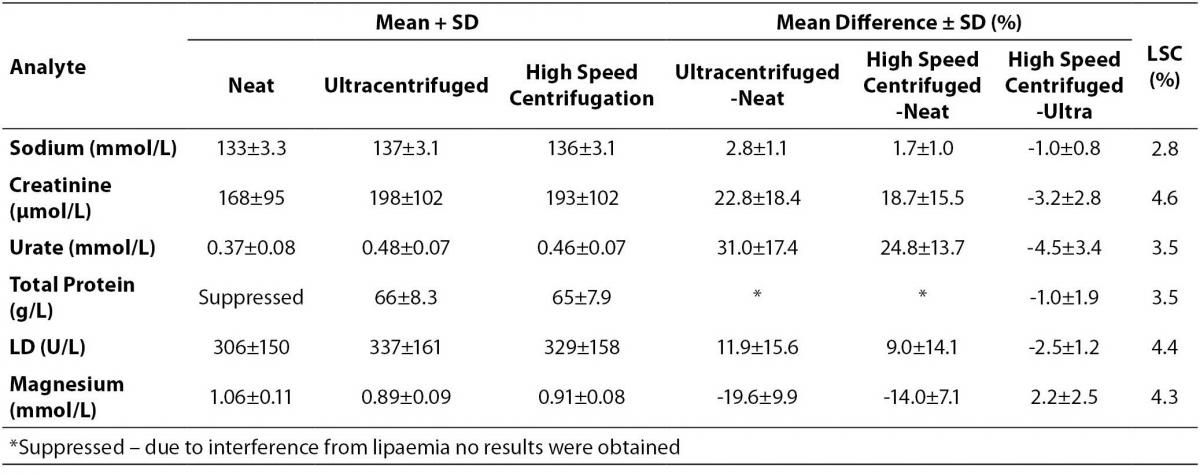
Table 2. Results of analytes affected by lipaemia compared between neat, ultracentrifuged and high speed centrifuged aliquots.
Apart from the urate result, all other results from the tested analytes indicate the difference between the high speed centrifuged and ultracentrifuged aliquots were within the LSC limits (Table 2). The total protein results in the neat sample aliquots were suppressed, no result was obtained due to interference.
Discussion
The data indicates < 1% of samples requested for lipid studies contain high enough triglyceride concentration (> 10 mmol/L) to potentially cause interferences in laboratory assays. When compared with the total number of samples received in the laboratory for biochemical analysis this represented one per ~4100 for the 2009-2010 period. For this study even with the aid of several other large laboratories outside our network it took more than a year to accumulate sufficient volume of fresh samples to prepare the 10 pools.
The lipaemic index along with the cholesterol and triglyceride levels data in Table 1 indicates the high speed centrifugation process evaluated here is effective in lowering lipid levels to allow sample analysis and produce results that are within LSC limits as confirmed in Table 2. Although the urate difference is outside the LSC limit, it is not considered clinically significant, < 10% difference (21). The effectiveness of the process in obtaining the clearest sample will also depend on the lipid levels and technique of aspirating the infranatant. For samples with higher lipid levels (e.g. triglyceride levels > 50.0 mmol/L) 3 or 4 centrifugations may be necessary provided there is sufficient sample available. Perhaps a request to the clinical unit to collect more blood tubes may overcome such a problem. This high speed centrifugation procedure appears to be superior to the one utilized by Vemeer et al (20), and this can be attributed to the much higher centrifugation speed and the double centrifugation process used in our study. A limitation of the study is not having performed a wider range of analytes due to insufficient sample volume (e.g. immunoassays) and suitable sample type (e.g. coagulation tests).
The Beckman DxC800 analyser measurable lipaemic index range is 0-10. Three of the pools with triglyceride levels > 30.0 mmol/L exceeded the upper limit, which made the lipaemic index less suitable than the triglyceride level for lipaemic interference studies. Review of the Beckman individual method inserts provides interesting information. For sodium, urate and LD the lipid interference studies used Intralipid concentration up to 500 mg/dL (in terms of triglycerides this equates to ~5.6 mmol/L), and it is reported there was no significant impact on any of these analytes. Our data showed these analytes are affected when the triglyceride concentration increases. For creatinine, it is stated lipaemic index up to 8 produced no significant impact, whereas with magnesium Intralipid concentration of 150 mg/dL produced an increase of ~0.16 mmol/L, and for total protein lipaemic index of 4 produced a decrease by 0.4 g/L, which is in agreement with our findings. Simply there is no standardized process with the lipaemic interference studies, and no triglyceride levels are provided for more direct comparison with our findings.
Native lipaemic samples in hospital environments most often originate from emergency departments, diabetic endocrinology and gastrointestinal units, lipid clinics etc which often require results urgently. To minimise delays in turn-around times of critical analytes such as sodium we analyse lipaemic samples first on blood gas analysers (bench top or portable) and report the electrolytes while the sample is ultracentrifuged. Most laboratories rely on the lipaemic index and manufacturer method recommendations for acceptable limits that are almost always established by using emulsions spiked samples in performing interference studies to determine if a sample will undergo ultracentrifugation or treatment. Others rely on measuring the triglycerides level, the most abundant lipid component in lipaemic samples before making a decision as how the sample will be processed because emulsions do not behave in the same way as native lipaemic samples. Many other laboratories use a combination of the two plus visually inspect the turbidity level, and this may be impractical in laboratories with large numbers of samples. It should be pointed that different analytical methods use different reagents and wavelength parameters, and will be affected differently by lipaemia levels and other non-lipaemic causes which includes lipaemic index methods. For example in most samples with triglyceride concentration > 11.0 mmol/L total protein on the Beckman DxC800 analysers will fail to produce a result. The Beckman lipaemic index method has been reported to produce high values due to non-lipaemic causes, precipitation of paraproteins (IgM kappa and lambda) in the Beckman serum index diluent (22). This requires samples with elevated lipaemic levels to be visually inspected for true lipaemia before reporting any results including the lipaemic index. In our experience this false high lipaemic index values in samples that are not visibly lipaemic can be correctly obtained by the use of normal saline as the index diluent.
Our practice with lipaemic samples presenting for the first time is to have lipids automatically performed even if they have not been requested by agreement with the hospital as a proactive service to aid clinicians with accuracy in patient care decision. Such lipaemic samples are indicative of serious and clinically important patho-physiological changes. This is now a standard procedure throughout our entire network of 33 laboratories servicing all the public hospitals in our state. Results reported from ultracentrifuged or lipid reduced samples are appropriately annotated to ensure clinicians are aware highest quality results have been reported. However, it is not advocated every laboratory implement such steps until it is authorized by laboratory management and clinical governance bodies of their hospital or health care institution.
One final point is the high speed centrifuges are not recommended for centrifugation of whole blood samples but serum or plasma only. Ideally laboratories will need to validate this procedure with there respective micro-centrifuges.
Conclusion
Not every laboratory can afford or will be equipped with an ultracentrifuge for the very small number of lipaemic samples that may be received per year. The procedure utilized in this study using a high speed micro-centrifuge showed it is effective in reducing lipid levels and produce a suitable alternative to ultracentrifuged samples to provide accurate analytical results. This procedure may serve as a standardized guideline in laboratories with high speed micro-centrifuges to improve the handling of lipaemic samples and minimise analytical errors.
Acknowledgement
We thank Robert Flatman and his staff, Sullivan and Nicolaides Pathology, Brisbane and Barbara Mottram and her staff, Mater Pathology Services , Brisbane for supplying lipaemic samples.
Notes
Potential conflict of interest
None declared.
References
1. Lippi G, Simundic AM, Mattiuzzi C. Overview on patient safety in healthcare and laboratory diagnostics. Biochem Med 2010;20:131-42.
2. Boone DJ. Is it safe to have a laboratory test? Accred Qual Assur 2004;10:5-9.
3. Muller MM. Quality and diagnostic perspective in laboratory diagnostics. Biochem Med 2010;20:144-6.
4. Carraro P, Plebani M. Errors in a Stat Lab: types and frequency 10 years later. Clin Chem 2007;53:1338-42.
5. Szecsi PB, Odum L. Error tracking in a clinical biochemistry laboratory. Clin Chem Lab Med 2009;47:1253-7.
6. Dimeski G. A commentary on the effect of lipid emulsions on pathology tests. Anaesthesia 2009;64:1033-5.
7. Goldenberg NM, Wang P, Glueck CJ. An observational study of severe hypertriglyceridemia, hypertriglyceridemic acute pancreatitis, and failure of triglyceride-lowering therapy when estrogens are given to women with and without familial hypertriglyceridemia. Clin Chim Acta 2003;332:11-9.
8. Lim KH, Lian WB, Yeo CL. Does visual turbidity correlate with serum triglyceride levels in babies on total parenteral nutrition? Ann Acad Med Singapore. 2006;35:790-3.
9. Drover JW, Cahill NE, Kutsogiannis J, Pagliarello G, Wischmeyer P, Wang M, Day AG, Heyland DK. Nutrition therapy for the critically ill surgical patient: we need to do better! JPEN J Parenter Enteral Nutr 2010;34:644-52.
10. Picard J, Harrop-Griffiths W. Lipid emulsion to treat drug overdose: past, present and future. Anaesthesia. 2009;64:119-21.
11. Kroll MH. Evaluating interference caused by lipemia. Clin Chem 2004;50:1968-9.
12. Dimeski G, Mollee P, Carter A. Increased lipid concentration is associated with increased hemolysis. Clin Chem 2005;51:2425.
13. Férézou J, Gulik A, Domingo N, Milliat F, Dedieu JC, Dunel-Erb S, et al. Intralipid 10%: physicochemical characterization. Nutrition 2001;17:930-3.
14. Twomey PJ, Don-Wauchope AC, McCullough D. Unreliability of triglyceride measurement to predict turbidity induced interference. J Clin Pathol 2003;56:861-2.
15. Bornhorst JA, Roberts RF, Roberts WL. Assay-specific differences in lipemic interference in native and intralipid-supplemented samples. Clin Chem 2004;50:2197-201.
16. Dimeski G. Interference testing. Clin Biochem Rev 2008;29 Suppl 1:S43-8.
17. Arambarri M, Oriol A, Sancho JM, Roncalés FJ, Galán A, Galimany R. Interference in blood coagulation tests on lipemic plasma. Correction using n-hexane clearing. Sangre (Barc) 1998;43:13-9.
18. Dimeski G, Mollee P, Carter A. Effects of hyperlipidemia on plasma sodium, potassium, and chloride measurements by an indirect ion-selective electrode measuring system. Clin Chem 2006;52:155-6.
19. McEnroe RJ, Burritt MF, Powers DM, Rheinheimer DW. Interference testing in Clinical Chemistry; Approved Guideline – Second Edition, CSLI document EP7-A2 (ISBN 1-56238-584-4). Clinical and Laboratory Standards Institute, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898 USA, 2005.
20. Vermeer HJ, Steen G, Naus AJ, Goevaerts B, Agricola PT, Schoenmakers CH. Correction of patient results for Beckman Coulter LX-20 assays affected by interference due to hemoglobin, bilirubin or lipids: a practical approach. Clin Chem Lab Med 2007;45:114-9.
21. Tanner M, Kent N, Smith B, Fletcher S, Lewer M. Stability of common biochemical analytes in serum gel tubes subjected to various storage temperatures and times pre-centrifugation. Ann Clin Biochem 2008;45:375-9.
22. Monk C, Wallage M, Wassell J, Whiteway A, James J, Beetham R. A monoclonal protein identified by an anomalous lipaemia index. Ann Clin Biochem 2009;46:250-2.